Abstract
Biomolecular interactions underpin almost all biological processes, and their rational design is central to programming new biological functions. Generative AI models have emerged as powerful tools for molecular design, yet most remain specialized for individual molecular types and lack fine-grained control over interaction details. Here we present ODesign, an all-atom generative world model for all-to-all biomolecular interaction design. ODesign allows scientists to specify epitopes on arbitrary targets and generate diverse classes of binding partners with fine-grained control. Across entity-, token-, and atom-level benchmarks in the protein modality, ODesign demonstrates superior controllability and performance to modality-specific baselines. Extending beyond proteins, it generalizes to nucleic acid and small-molecule design, enabling interaction types such as protein-binding RNA/DNA and RNA/DNA-binding ligands that were previously inaccessible. By unifying multimodal biomolecular interactions within a single generative framework, ODesign moves toward a general-purpose molecular world model capable of programmable design.
Highlights
- First-of-its-kind all-to-all world model: Unified framework enables cross-modality molecular generation—proteins, nucleic acids, and small molecules—within a single architecture built upon AlphaFold3-like structure prediction.
- Multi-level controllable generation: Task-oriented masking mechanism provides fine-grained conditional control at entity-, token-, and atom-levels, supporting binder design, motif scaffolding, and atomic motif engineering.
- Flexible and rigid design modes: Supports both fixed-target (rigid receptor) and co-design (flexible receptor) strategies, enabling epitope-specific generation through hotspot residue guidance.
- Superior performance and throughput: Consistently outperforms modality-specific baselines across 11 benchmark tasks, achieving 2-4 orders of magnitude improvement in design throughput with minutes per sample.
- Previously inaccessible interactions: Enables novel design capabilities including protein-binding RNA/DNA aptamers and nucleic acid-binding small molecules that were computationally infeasible before.
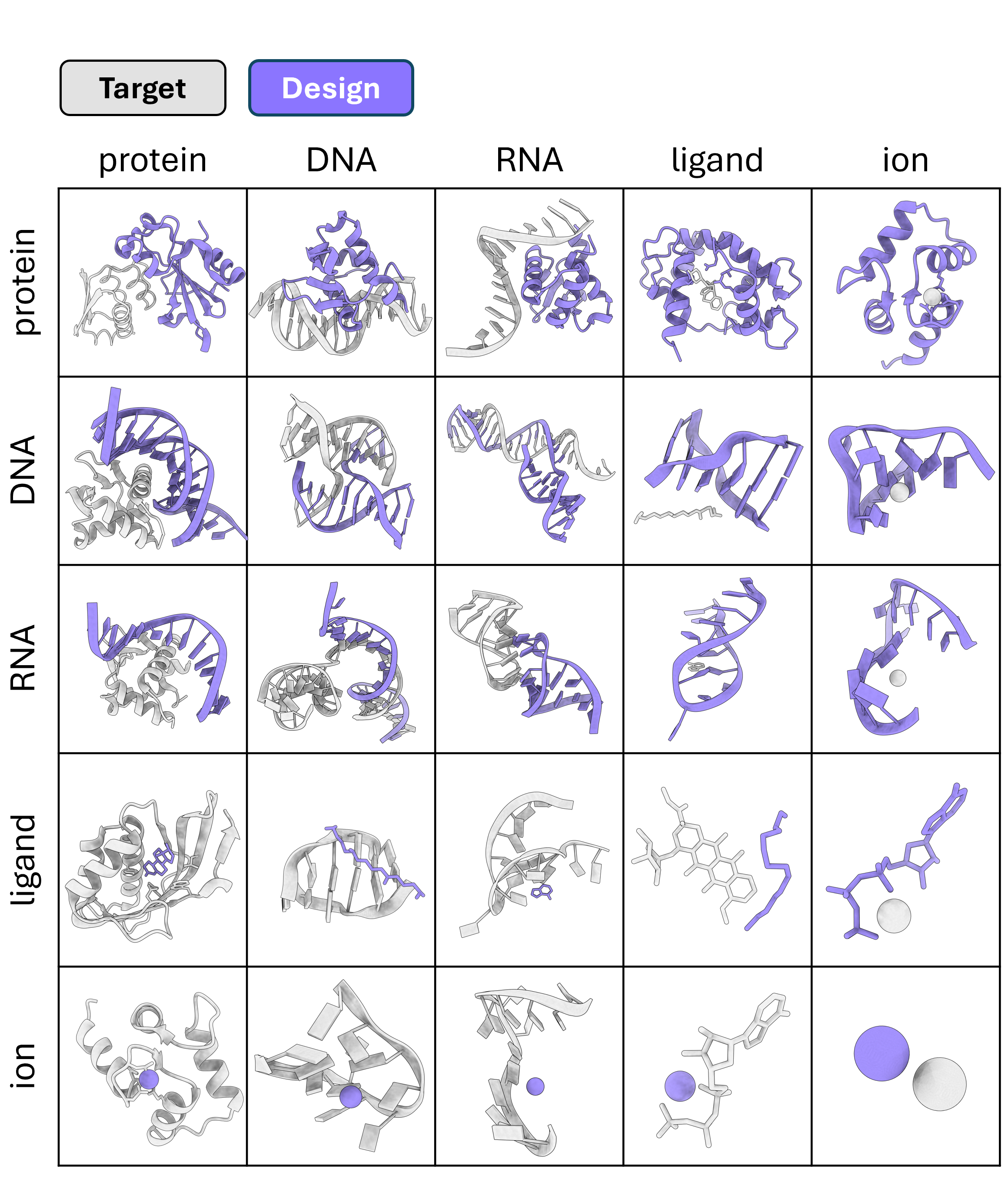
ODesign is an All-to-All design framework that simultaneously models targets (grey) and designed structures (violet) across proteins, nucleic acids, and ligand complexes.
Design cases
Showcasing the dynamic design process of protein, cyclic peptides, nucleic acids and ligand molecules.
Architecture
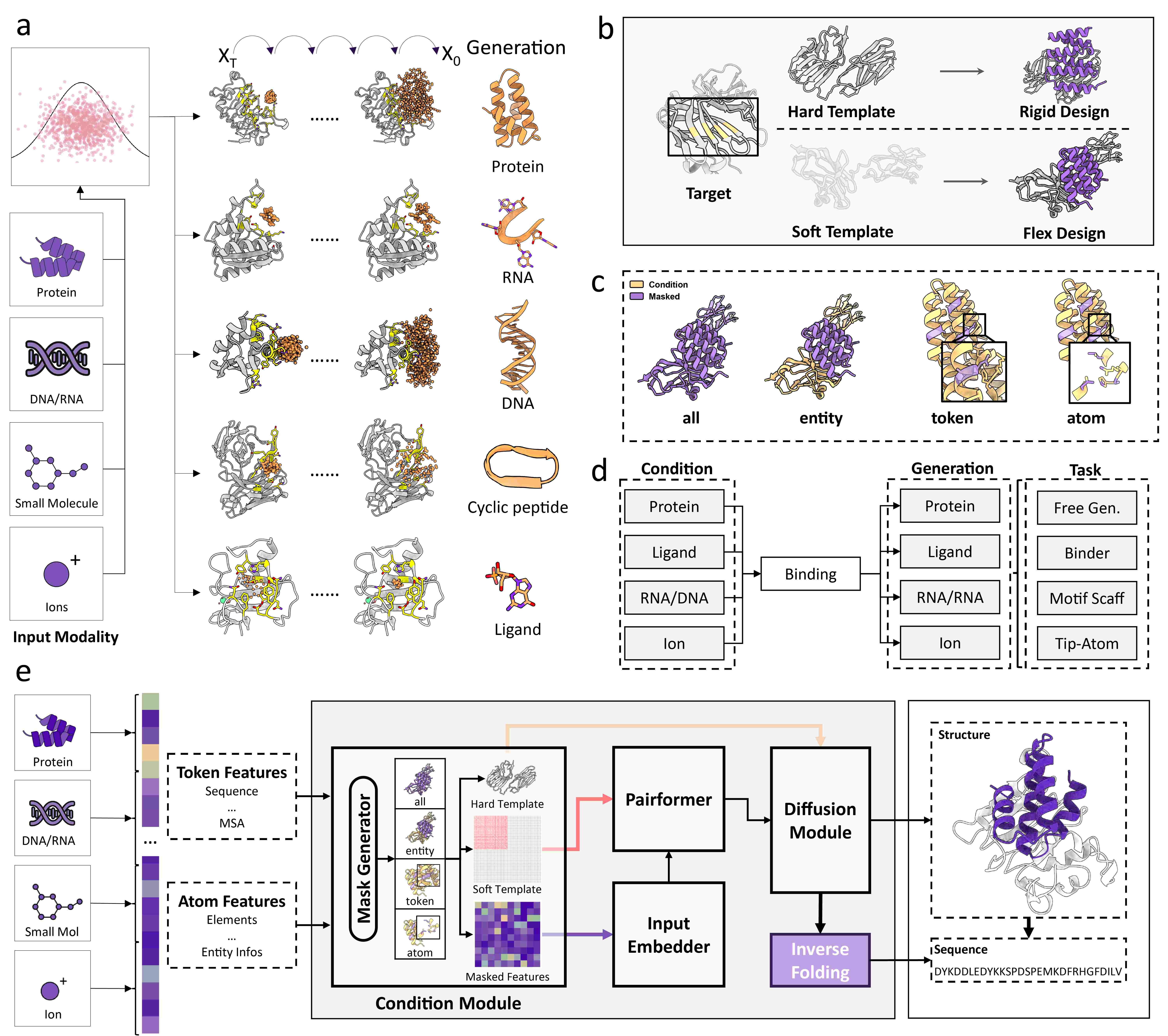
1. Embedding Module
Unified generative tokens abstract minimal chemical units across modalities. Target 3D structures are incorporated as initial coordinates and distance constraints.
2. Conditional Module
Dual control mechanism: initial coordinate "guess" and distance-based interaction modeling. Hotspot residue features enable epitope-specific design.
3. OInvFold Module
Unified inverse folding assigns sequences to protein/nucleic acid backbones and atom types to ligand scaffolds with modality-specific decoders.
In-silico Performance
ODesign consistently outperforms modality-specific baselines across protein, nucleic acid, and ligand design benchmarks.
Protein Design
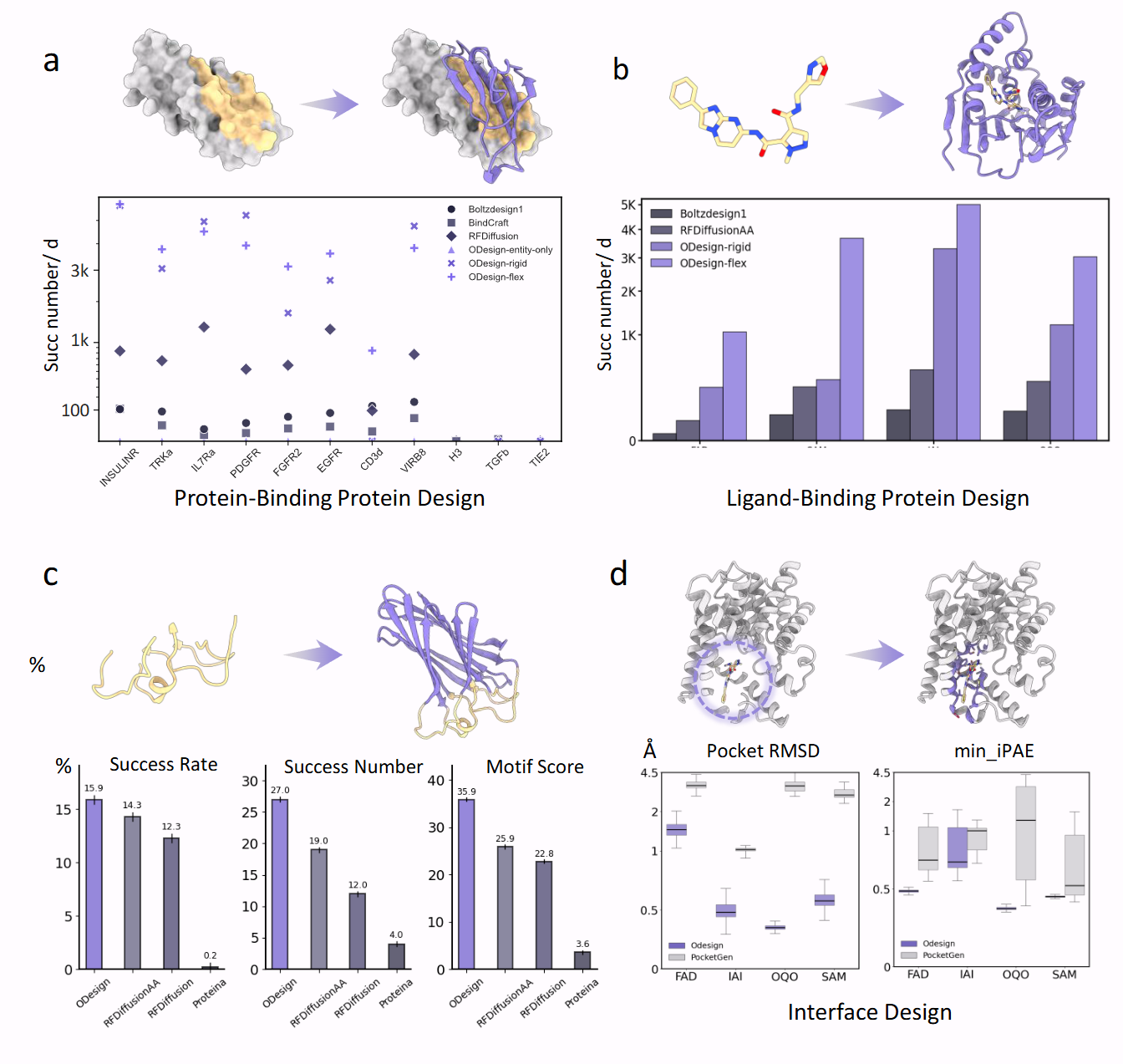
Performance on protein-centric benchmarks: binder design, ligand-binding protein design, motif scaffolding, interface design, and atomic motif scaffolding
Nucleic Acid Design
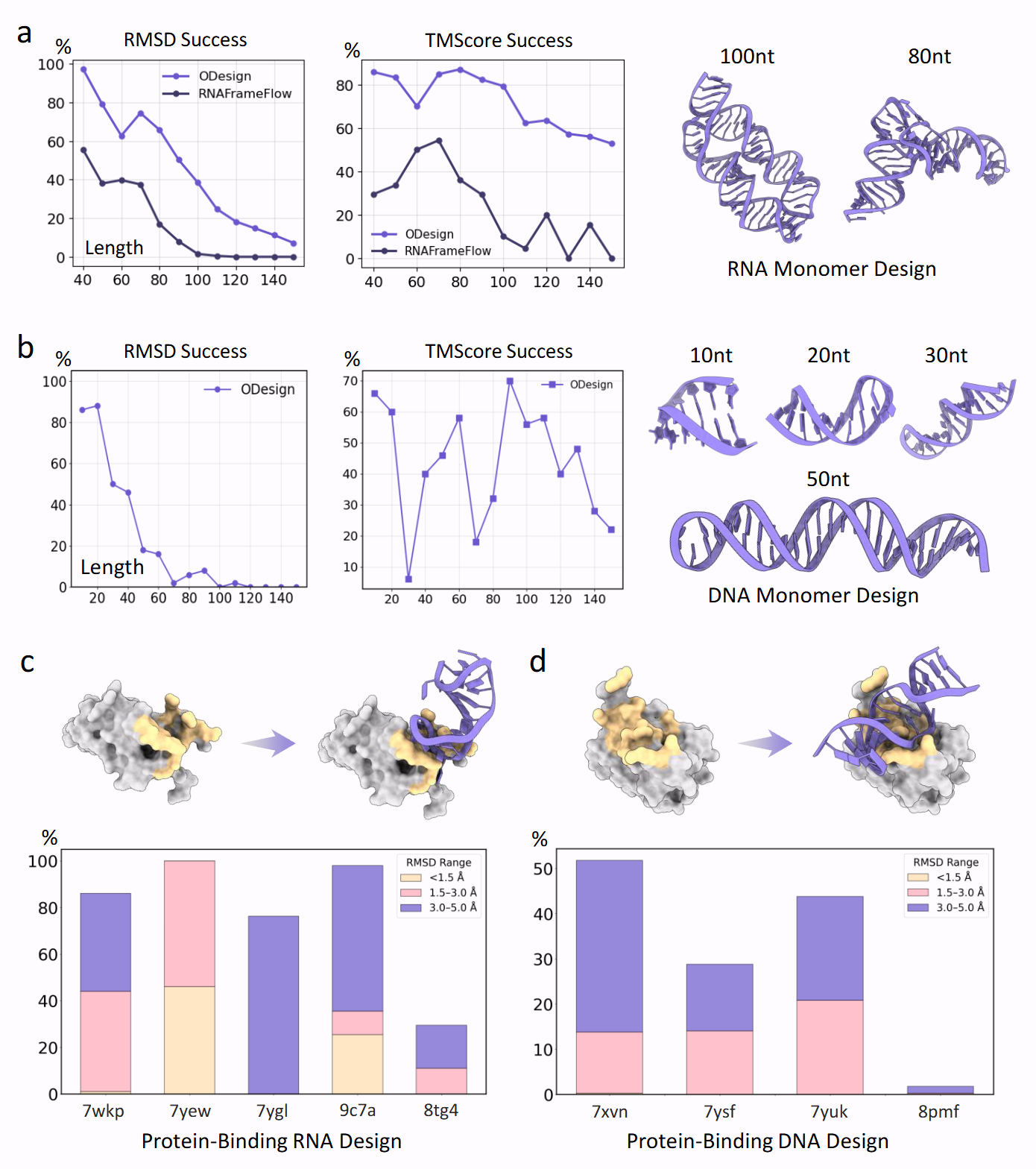
Performance on nucleic acid benchmarks: RNA/DNA monomer design and protein-binding RNA/DNA aptamer design
Ligand Design
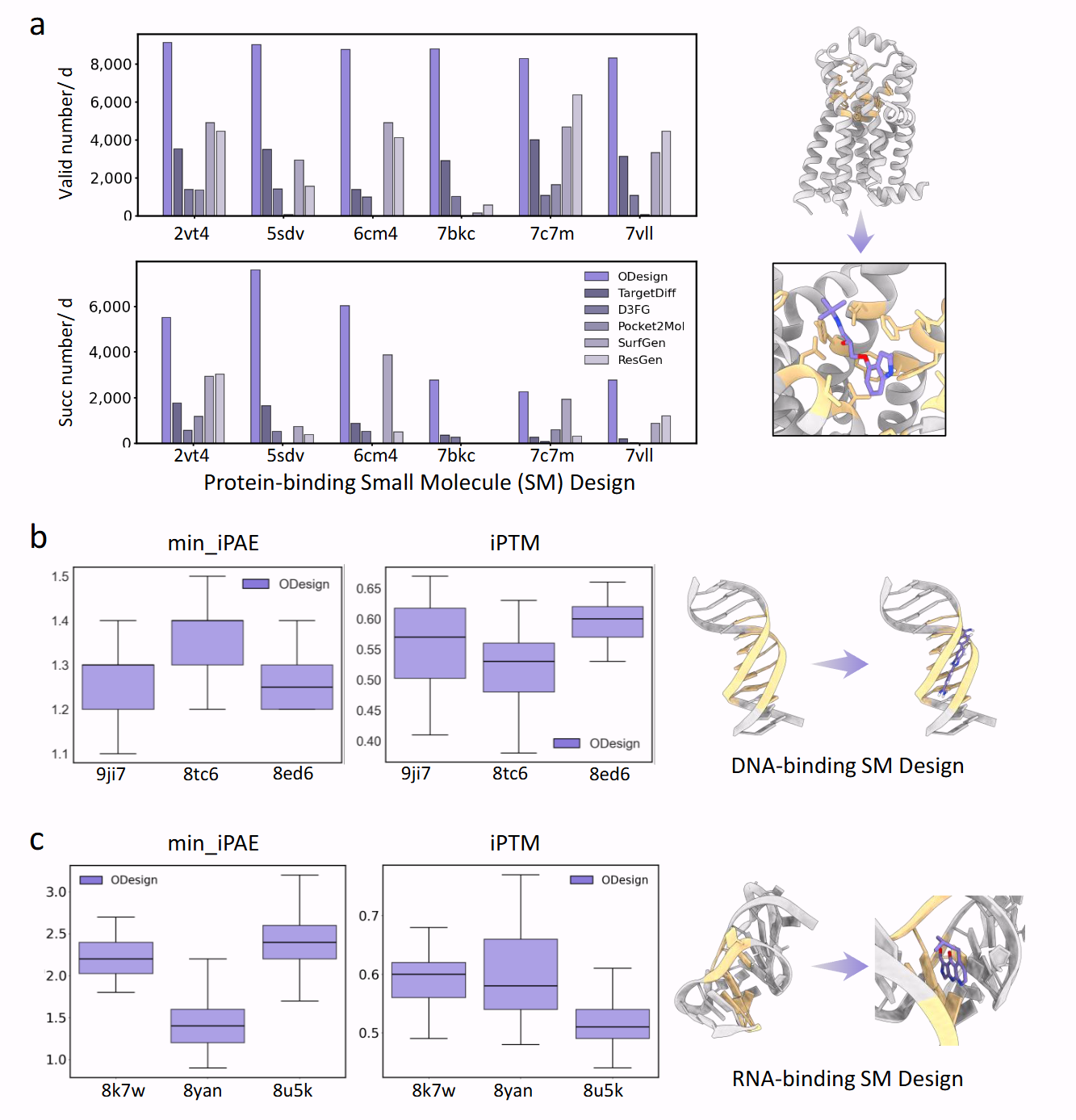
Performance on ligand-centric benchmarks: protein-binding, DNA-binding, and RNA-binding small molecule design
Contact Us

Server


Friendly Link: Odin Zhang If the ODesign server group is full, please contact me on wechat: zhanghaotianzds